
 |
 The functionality described in this
topic is only available when you mark Show
Advanced Options.
The functionality described in this
topic is only available when you mark Show
Advanced Options.
Add/Review Content
Create gRNAs
When you select to add content using the Create gRNAs method, you first define the targets that you want to capture (screen 1), then you review the targets to make sure that SureDesign successfully recognized all your targets (screen 2). Finally, you assign the selection parameters using a set of drop-down lists (screen 3).
When you are finished making your selections, you then submit the design to SureDesign and the program's algorithms select the gRNAs. You receive an e-mail from Agilent SureDesign notifying you when your gRNA selection job is complete and the results are available for you to review.
In this step, define the DNA regions of interest for the gRNAs (referred to in the wizard as a "targets") by providing the following information:
Targets
In the Targets text area, enter one or more identifiers for the target using either of the following approaches:
· Type or paste the target identifiers.
· Click Upload to browse to a text file (*.txt) that contains the target IDs.
The permitted identifiers are:
· For target genes:
Gene name - enter the gene name (not case-sensitive) as it appears in one or more of the selected databases; example: brca1; see SureDesign gene finder for information on how SureDesign maps a gene name to a specific genomic location
Transcript ID - enter the transcript ID (not case-sensitive) as it appears in one or more of the selected databases; examples: NM_007294, OTTHUMT00000348798, or ENST00000357654; note that SureDesign ignores version numbers included in the transcript ID
Gene ID - enter the numerical NCBI gene ID; example: 672
GO ID - enter the GO Id; example: GO:0048040
· For target genomic intervals:
Genomic coordinates - enter the chromosome number and range of nucleotides using the UCSC browser format or BED format.
You can add a string of text, no spaces, after the target genomic interval to be used as the target ID (e.g. chr1:1-100 geneX).
Databases
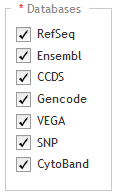
Below the Databases heading, mark the genome annotation databases that you want SureDesign to use to obtain genomic coordinate information for your specified targets. The databases that you have to choose from are dependent on the species you selected in the Define Design step. For H. sapiens, the available database sources are:
RefSeq - US National Center for Biotechnology Information (NCBI)
Ensembl - European Bioinformatics Institute and the Wellcome Trust Sanger Institute
CCDS - Consensus Coding Sequence project (CCDS) of the US National Center for Biotechnology Information (NCBI)
Gencode - US National Human Genome Research Institute (NHGRI) and the Wellcome Trust Sanger Institute
VEGA - Vertebrate Genome Annotation project of the Human and Vertebrate Analysis and Annotation (HAVANA) group at the Wellcome Trust Sanger Institute
SNP - dbSNP database from the National Institutes of Health (NIH)
CytoBand - CytoBand file from the UCSC Genome Browser
Regions of Interest
Specify the specific regions within the targets for which you want to select gRNAs. Use the options below the Regions of Interest heading:
· Entire Transcribed Region - Select this option to include gRNAs for the entire genomic sequence (exons, introns, and UTRs) of your target genes.
· Coding Exons - Select this option to include gRNAs only for the translated regions of the target genes. If you want to include only the first coding exon and exclude all other coding exons, make sure the check box labeled First Exon is marked. (If the First Exon check is not marked, then none of the coding exons for the target genes are excluded).
· Transcription Start Site - In case of Transcription Start Site (TSS), gene finder will provide the 1st base from where transcription begins. Genes may contain multiple TSS depending on the number of transcripts. Start site will also depend on the orientation of the transcript.
NOTE For target genomic intervals (i.e. targets entered as genomic coordinates), SureDesign always includes the entire genomic sequence when selecting sequences for the design, regardless of your selection for the Regions of Interest.
Include Flanking Bases
In the 3' and 5' drop-down lists, select how many base pairs of flanking sequence (on the 3' and 5' ends, respectively) you want SureDesign to include with the target region when designing the gRNAs.
NOTE SureDesign does not include flanking bases for targets entered as genomic coordinates.
Allow Synonyms
When this check box is marked, SureDesign compares the gene names you entered into the Targets area to a table of synonyms, and may use the synonym names to map the genes to a genomic location. For example, if you entered HER2 as a target, SureDesign would identify HER2 as a product of the gene ERBB2, and use ERBB2 to map the genomic location.
In cases in which the gene name for your target is also a synonym for another gene, SureDesign treats both genes as targets when Allow Synonyms is marked. For example, if you entered DSP as a target, SureDesign would identify your target as the official gene name for desmoplakin, but it would also identify it as a synonym for the gene encoding dentin sialophosphoprotein. Consequently, the program would map the genomic location to two completely different genes, and in the next step of the wizard (Step 3: Review Targets), you would see both genomic locations listed for the target.
When the Allow Synonyms check box is cleared, SureDesign maps your targets to genomic locations using only the entered gene names.
To fully control how SureDesign maps your targets to a genomic location, enter your targets using transcript IDs, gene IDs, or SNP IDs instead of gene names. Alternatively, after you advance to the Review Targets step of the wizard, click Download to download the Regions.bed file and then edit the genomic locations listed in the file so that they accurately match those of your targets. You can then go back to the Define Targets step of the wizard and paste the genomic locations into the Targets input area.
Click Next to continue to Screen 2.
This screen provides a chance for you to make sure that SureDesign successfully recognized all of the target identifiers that you entered on the previous screen. Review the Target Summary and Target Details before you click Next.
Target Summary
Near the top of the wizard window is a target summary with two bullet points that indicate:
· 1st bullet point: The number of targets that SureDesign was able to resolve to a genomic location, and the total number of continuous genomic regions that comprise those targets.
· 2nd bullet point: The number of targets that SureDesign was not able to find in any of the databases you selected on the previous screen.
 If SureDesign did not accurately identify
all of your target regions
If SureDesign did not accurately identify
all of your target regions
The Target Details table lists the following information for each of the target identifiers that SureDesign was able to locate:
· Target - The Target column lists the gene name, transcript ID, SNP ID, or genomic coordinates that you used to define the target.
· # Regions - The # Regions column lists the number of target regions within the target.
· Base Pairs - The Base Pairs column lists the total number of base pairs within the regions defined by the target identifier.
· Position - The Position column lists the genomic coordinates identified for the target.
· Group IDs - The ID(s) used to define the target (e.g., GO or KEGG ID).
NOTE If you entered the target sequence using a gene name, accession number, or similar identifier, click View targets in UCSC to open the UCSC Genome Browser and see the genomic location of the target identified by SureDesign.
Click Next to advance to Screen
3.
At this step, enter the parameters for the gRNA selection process. When you are finished making your selections, submit the design to SureDesign to begin gRNA selection.
Filters
· PAM Sequence - Select which PAM sequence you want SureDesign to use when selecting gRNAs for the target window. You can select between NGG and NGGNG, or select Custom to enter a PAM sequence of your choice. If you select Custom, a field appears next to the PAM Sequence drop-down list where you can enter the desired custom PAM sequence.
· Search In Strand - Select which strand of the target window sequence you want SureDesign to use when selecting gRNAs. You can select Forward (for the + strand), Reverse (for the – strand), or Both (for both + and - strands).
· gRNA with GG at the 5' end - Mark this check box if you want SureDesign to only select gRNAs that have an endogenous GG at the 5' end of their sequence.
Scoring Algorithm
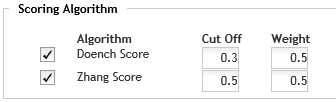
Select one or both of the available scoring algorithms.
· Doench Score - Calculates an on-target score using the method described in Doench, et. al. (Nature Biotechnology 34, 184-191 [2016]).
· Zhang Score - Calculates an off-target score using the method described in Zhang, et. al. (Nature Biotechnology 31, 827-832 [2013]).
Set the parameters for the selection algorithms.
· Cut Off - This parameter sets the minimum score threshold for the gRNA sequences. gRNAs are discarded if their score is below the specified cut off. You can enter a value from 0.00 to 1.00. Note that each scoring algorithm has its own Cut Off parameter.
· Weight - The Weight parameters set
the relative weights of the two scoring algorithms, which SureDesign then
uses when calculating the final scores for the gRNAs. For example, if
both algorithms are selected, and both weights are set to the default
value of 0.5, then the two algorithms are equally weighted when calculating
the final score for a gRNA. The formula used for the final weighted score
is:
(WeightDoench
× ScoreDoench)
+ (WeightZhang
× ScoreZhang)
To
submit the design for gRNA searching:
When you are finished entering the parameters, submit the design to the SureDesign job queue and the SureDesign algorithms will search for gRNA sequences for your design.
Click Find
gRNAs.
A message box opens indicating the e-mail address where Agilent will
contact you when the probe selection job is complete. If desired,
you can enter additional e-mail addresses into the provided field.
Click OK
in the notification message to submit the design to SureDesign.
Your submission is placed in the SureDesign job queue.